Reading the Internet Is a Medical Device: The FDA and 23andMe

By MATT SHAPIRO
A few years after shutting down 23andMe’s genetic health reports, the FDA and 23andMe have come to an agreement on what health related information can be delivered to 23andMe customers. I wrote about the new health reports in detail (and will write about them again when I get my profile updated to include these reports), but they are still a far cry from the health reports 23andMe used to give out.
When I first heard about the FDA shutting down 23andMe’s health report, I was deeply upset. Here, I thought, was a great company, delivering important health information to their customers at a staggeringly low price. Not only that, but they were also delivering cutting-edge genetic health information for the same price that a doctor charges to put you on a scale, measure your weight and height, and listen to you breathe for 2 minutes. As health information goes, the 23andMe genetic health report was a thousand times more valuable than a check-up and cheaper to boot.
Then I heard people make the case that 23andMe was basically ignoring the FDA’s perfectly reasonable pleas that they subject themselves to federal regulation because telling people what medical conditions their DNA pre-disposes them toward should count as a diagnostic test.
Having dug deep into my own personal genome and exploring the world of data and personal genetics, I came to believe that the FDA’s position was wrong on grounds of the basics of human autonomy and our right and freedom to know about our own bodies. But that is largely because I subscribe to a culture very different from the world of medical regulation.
What 23andMe Used to Tell You
I do not have first-hand access to the old 23andMe health reports since I had my genome sequenced after the FDA shut down the health reports. Instead, I have to draw from old 23andMe blog posts on the reports they delivered and how to read them.
The results of most medical studies involving correlations are put forward as an identification of risk. The recent “processed meat causes cancer” report is a near-perfect example of that. Researchers find that, in a number of studies across a range of population groups, there is a positive correlation between diets high in processed meat and higher-than-normal rates of colorectal cancer. Many studies have been done on this correlation and it is fairly consistent.
However, the rate of colorectal cancer is only elevated 18 percent for high meat diets in comparison to no-meat diets. So out of 200 people with high-meat diets, 12 of them will contract colorectal cancer compared with 11 people with no-meat diets. So, does meat cause cancer? Yes. But only when we look at a large enough population. It probably won’t cause cancer in you. But you could also be very unlucky.
This is how the results of nearly every genetic study work. They don’t say “You have this gene, therefore you will suffer atrial fibrillation.” Instead they say “People who have this gene are 41 percent more likely to have atrial fibrillation in the course of their lifetime.” And that bumps up your risk from 15.9 percent (without the gene) to 22.4 percent (with the gene).
I’ve worked as a developer and designer for medical software and one of the greatest challenges is trying to find ways to make medical information readable and understandable. Very few things are harder to communicate than the nature of risk. This was the challenge that 23andMe faced when they first developed their health reports. It’s worth noting that this was pre-FDA approval. No one forced them to help their users understand genetic risk in a valuable, accurate way. They did it because they wanted to.
In order to help their customers understand the results of the health reports, 23andMe had to convey information about
- confidence in the results – is it one study that showed a small correlation between a genetic marker and elevated disease risk? Or several studies that showed strong correlations?
- magnitude of the relative risk – how much does a given genetic marker increase your risk relative to the general population? Twice the risk? Four times?
- magnitude of the absolute risk – you could have a genetic marker that substantially elevates your relative risk, but maybe it is for a condition that is rare in the same way that buying 10 lottery tickets instead of one dramatically increases your risk of being hilariously rich.
- mixed risk scenarios – for some well studied diseases, there are multiple markers across several chromosomes that have been found to correlate with that disease. Sometimes people have a mixture of increased-risk marker and decreased-risk markers. How do you communicate something that complex?
From what I’ve seen, 23andMe managed to deliver a report that gave an enormous amount of information and did a pretty good job of communicating that information to their customers. Just to look at a few of their samples:
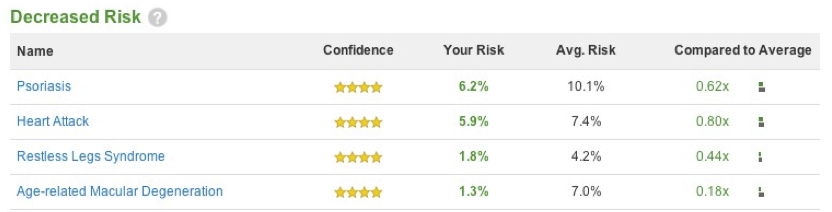

In these elevated and decreased risk reports, they appropriately focus on the high-confidence markers (requirement 1). In fact, they actually didn’t give the user any information on the front page of the report for anything that wasn’t high confidence. They gave their users the relative risk (requirement 2 in the “Compared to Average” column) and the ability to compare absolute risks (requirement 3) in the “Your Risk / Avg. Risk” columns. They provide a bar chart to the left to try to help users grasp the nature of the absolute and relative risks together, although they have so little space to show that information I probably would argue that it isn’t incredibly helpful.
If we’re talking about visualization, I would say that the following screen is a much better way of trying to communicate absolute and relative risk.

Here, the designers have offered an excellent visual to communicate their sample user’s genetic Type 2 Diabetes risk. This screen could be seen when the user clicks on a report (I assume) which means that the user indicated their desire to learn more about this specific risk.
I also like 23andMe’s decision to *not* show information about lower-confidence risk results. Instead of having their users freak out over a mixed risk result, they require user interaction to examine the results and encourage the user to do an informational deep-dive into markers and traits that are important, but complicated.

This is a huge, all-encompassing, health report on a complicated subject. Understanding it requires customers to educate themselves and understand the nature of risk reports. In short, the person who will understand this report the most and get the most out of it is the person who is the most literate in modern science.
However, if you dig through the 23andMe blog or sign up for their test, you’ll find that 23andMe didn’t just hand people these reports and say “you’re on your own.” I would go so far as to say that 23andMe had two missions: the first was to do personal genetics testing and deliver the results to their customers and the second was to educate their customers on the results that they got. 23andMe took the responsibility to educate their customers very seriously and devoted a lot of time and money to this end.
How Did 23andMe Get This Information?
When I did my 23andMe genetic testing, they were not offering health reports. I was not overwhelmingly optimistic that they would bring back their health reports (and in a way I was right) but I didn’t care because I love data and the idea of having the granular data about who I am was overwhelmingly exciting to me.
23andMe gave me my specific alleles for more than 600,000 SNPs. Let me decode that:
An SNP (single nucleotide polymorphism) is a specific spot in your DNA. For example: The DNA string of your 15th chromosome is 102 million base pairs long. At position 78,601,997, researchers have identified a place where different people have different combinations. These specific points in our DNA are called SNPs and the different combinations we might have are called “alleles.” If your DNA at that spot, your allele, is C:C, that is considered normal. If you have the C:T, you’re at 30 percent higher risk for lung cancer. If you have T:T, you’re at 80 percent higher risk for lung cancer.
(If this is still way too confusing, check out my other over-long piece on the basics of understanding genetic data.)
An increased risk of 80 percent for lung cancer sounds bad, but let’s put this in perspective: If you smoke cigarettes, you are increasing your risk for lung cancer by 1500 to 3000 percent. Smoking is linked to 90 percent of lung cancers. That is huge. Compared to smoking, the rs1051730 SNP is a marginal player.
I know what my allele for rs2464196 because I can download my data from 23andMe. But if you are a 23andMe customer and you don’t have the time or inclination to download your raw data in bulk, you can also search for a specific SNP on their website.
That’s how I know my personal genetic marker for this risk factor … but how do I know the relative risk associated with that data?
Because the federal government told me.
Actually, that’s a bit of a simplification. Genome.gov is a website that hosts a huge amount of research through the US National Library of Medicine and is sponsored through the National Institute of Health*. They host papers that talk about the risks associated with rs2464196, specifically the lung cancer risk.
So I learned about my genetic risk through a government website. If I had no job and a truly heroic attention span, I could look up my SNPs one by one, maybe 60 to 100 per day. I could catalog the risk into a spreadsheet and, eventually, I would be able to generate a health report that is not unlike the one that 23andMe delivered to their users: a report based strictly on data that is publicly available, much of it through a government-hosted website.
Or I could write a script to crawl all this data, catalog it and run it against my personal genetic data, downloaded from 23andMe. This basically just out-sources the task of me reading the internet; I tell an app to read it for me and report back on what it finds. This is the beauty of programming: take a task that is tedious for humans, automate it and learn something new about the data.
To the best of my understanding, this is what 23andMe was doing. They were reading the internet, reading government websites filled with valuable genetic information, and reporting the results of that public information back to their users. Oh, sure, they’re probably augmenting their results with information that they, as a private company, have gleaned. But I suspect that most of their information comes from reading the open SNP information from open sites and reporting on the results to their users.
But if 23andMe is just automating the public process of reading the internet, why would the FDA shut them down?
Why the FDA Wasn’t OK With This
The difficulty the FDA had with 23andMe was that what 23andMe was doing was so revolutionary that it didn’t fit into any clean categories that the FDA had. It was a consumer product that was health-related, yes. Based on hard, public science, yes. But is it a diagnosis? A medical device? A health screening? It was utterly unlike anything the FDA was currently regulating.
When it comes to medical information in a consumer product, the FDA is concerned with two things:
- Is this information accurate?
- Can the consumer understand the information well enough to make health-related decisions based on it?
The answer to the first question is reasonably easy. The devices 23andMe uses to tell you your genetic results have defined error rates, and I assume 23andMe can do multiple “passes” to make sure the information you’re getting about your own DNA is accurate. We can put numbers to the “is this accurate” question.
We can’t, however, put numbers to the “can consumers understand this” question. Or, at least, it’s much harder to do so. 23andMe would need not only to educate but also to verify that their users were actually absorbing the education and not making rash decisions about their genetic information.
By all accounts, 23andMe didn’t really understand the nature of the FDA’s initial requests. They were looking at the regulatory questions through the eyes of technologists, essentially saying “Hey, we gave our customers the information they need to make informed decisions, and we’re doing our best to help them make good decisions, but ultimately it’s not our fault if they are given accurate information and jump to terrible conclusions.”
The villain in me says that if this is an inappropriate view of reporting health information, every blog and newspaper in the world should be shut down by the FDA since they routinely misreport and misunderstand scientific papers, encouraging their readers to jump to terrible conclusions. Terrible conclusions are an inevitable consequence of any kind of information sharing and we cannot, on this side of heaven, hope to eliminate that.
Still, there is this strange dichotomy in which the science is out there, hosted by the government, and available to anyone who cares to go look at it. And 23andMe was reading it to their customers along with a lot of helpful information above and beyond the basic scientific results. The government makes this information available, but you can’t sell a service that reads it without FDA approval.
[Seinfeld Voice] What Is the Deal With The FDA?
The problem the FDA had with reading these scientific papers to their consumers largely comes down to the fact that it’s hard to teach people to understand science. Science (especially genetic science) is a moving target and there is a lot of genetic information that 23andMe didn’t give to their users either because they didn’t know or because the science is constantly shifting. There are scenarios in which I might have a marker that increases my risk of cancer, but another marker that decreases it and 23andMe only reports on one of those markers.
Genetics health diagnosis is incredibly complicated and the case can be made that 23andMe simplifying it into a simple percentage of risk, while accurate for the individual marker, provided an inaccurate overall health assessment. Or, at the very least, an assessment that gave users a false sense of confidence in the results.
I could still get a false sense of confidence by reading the raw science myself, but this approach the FDA has taken is analogous to their approach with other medical devices.
Let’s say I had diabetes and I have an insulin pump. I could create an Arduino project that charts my blood sugar levels and automates the insulin delivery while I sleep. I could do this for myself and it is potentially life-saving … but also potentially life-threatening. But if I try to do it for someone else, it’s a medical device and subject to FDA regulations. Due to these regulations, I can’t even put the code on how to do it online because the code is considered a part of that medical device. I can play around with my own life and health, but the FDA draws the line where I try to impact other people’s lives.

While watching the 23andMe drama unfold, I got the feeling that the FDA is operating under an old paradigm of how to manage heath information in a way that protects consumers from themselves. As I’ve learned more and more about the inner working of this story, it sounds like the FDA was somewhat caught off guard by the explosion of consumer genomic sequencing and has been working to try to find the best ways to regulate this new and incredible way of looking at health.
The good news is that we’re in the middle of this story, not at the end. The FDA approval of 23andMe releasing limited health information will likely be soon followed by FDA approval of addition health information, slowly repopulating the amount of health information 23andMe is allowed to convey to their users. The difference is that this time 23andMe will be working carefully with the FDA to make sure that the information released meets the FDA’s threshold for accuracy, value and clarity to the customers.
I was initially worried that the legitimate concern with protecting consumers will hamstring organizations who are fighting hard to push forward to make the amazing progress that is possible. But this is more a story of two organizations with vastly different visions of the consumer. 23andMe was coming from a tech culture of moving fast, creating value, and making information free and accessible. The FDA culture is slow, cautious and deeply concerned with verifying everything 10 times over before approving.
My personal preference is to lean toward the tech view of doing things because that is where I come from. But in the ongoing conversations, it sounds as if both organizations are changing. 23andMe is slowing down and learning to provide the verification the FDA wants, while the FDA is learning to speed up so consumers can take advantage of the most exciting thing to happen to medicine since vaccine development.
*Full disclosure: the head of the NIH is Dr. Francis Collins, who is my personal genetics superhero. He can, in my fanboy mind, do no wrong.
Matt Shapiro is a software engineer, data vis designer, genetics data hobbiest, and technical educator based in Seattle. He tweets under @politicalmath, where he is occasionally right about some things.



